Improved dimer method: Difference between revisions
(Created page with "The dimer method<ref name="henkelman1999"/> is a technique for the optimization of transition states. In VASP, the method improved by Heyden et al.~\cite{Heyden:05} (IDM) is i...") |
No edit summary |
||
| (34 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
The dimer method | The dimer method{{Cite|henkelman:jpc:1999}} is a technique for the optimization of transition states. In VASP, the improved dimer method (IDM) by Heyden et al. is implemented, and a detailed presentation of the method can be found in their paper{{Cite|heyden:jpc:2005}}. | ||
In VASP, the method | |||
detailed presentation of the method can be found in | |||
The | The initial curvature along the dimer axis is computed using finite differences. The initial dimer direction must be provided (see below). The IDM procedure shown in Figure 1 is described in four steps with {{TAG|FINDIFF}} = 1: | ||
[[File:IDM.png|800px|thumb|left|Figure 1. The IDM is relaxed on the potential energy surface (PES) in four ionic steps. Solid arrows show the dimer axis '''u'''<sub>ξ</sub> and solid circles the structure for which the forces are calculated in the step. The empty circles and dashed lines indicate the structures and dimer axes from the previous steps. The dotted arrow in '''(d)''' represents the dimer axis on rotation by φ<sub>min</sub>.]] | |||
is | <div style="clear:both;"></div> | ||
'''a)''' An initial direction '''u'''<sub>ξ</sub> is taken from the most negative vibrational mode of the trial structure. The trial structure is the first point '''q'''. | |||
'''b)''' An additional point on the potential energy surface (PES) forward along the trial direction is defined '''q''' + δ'''u'''<sub>ξ</sub>. The first and second points together define the dimer. | |||
'''c)''' The dimer is then rotated on the PES about '''q''' by angle φ<sub>1</sub> to '''q̃''' such that its axis is parallel to the direction of maximal negative curvature. | |||
'''d)''' A new direction is defined by rotating '''u'''<sub>ξ</sub> by φ<sub>min</sub> to minimize the negative curvature of the PES λ. A search direction N̅ is defined using a minimization algorithm, then a translation (optimization step) is taken in the unstable direction of N̅ to '''q''' + εN̅, where ε is a step distance. The potential energy is is maximized along the unstable direction, (i.e., dimer axis) while it is minimized in all other directions. | |||
Rotation followed by translation is followed iteratively until convergence, i.e. the saddle point, is reached. | |||
The method is invoked by setting {{TAG|IBRION}}=44 in the {{TAG|INCAR}} file. | |||
Furthermore, the user must specify the direction of the unstable mode. The corresponding <math>3N</math> dimensional vector is defined in the {{FILE|POSCAR}} file after the lines with atomic coordinates and a separating blank line. Note that the dimer direction is automatically normalized, i.e., the norm of the dimer axis is irrelevant. An example of a {{FILE|POSCAR}} file for a simulation with the dimer method is given in the following: | |||
{ | |||
ammonia flipping | |||
1. | |||
6. 0. 0. | |||
0. 7. 0. | |||
0. 0. 8. | |||
H N | |||
3 1 | |||
cart | |||
-0.872954 0.000000 -0.504000 ! coordinates for atom 1 | |||
0.000000 0.000000 1.008000 | |||
0.872954 0.000000 -0.504000 | |||
0.000000 0.000000 0.000000 ! coordinates for atom N | |||
! here we define trial unstable direction: | |||
0.000001 0.522103 -0.000009 ! components for atom 1 | |||
-0.000006 0.530068 0.000000 | |||
-0.000005 0.522067 -0.000007 | |||
0.000001 -0.111442 0.000001 ! components for atom N | |||
As in the other structural optimization algorithms in VASP, convergence is controlled through the {{TAG|EDIFFG}} tag. | |||
{ | |||
\ | Experienced users can affect the performance of the dimer method by modifying the numerical values of the following parameters (the given example values are the default values): | ||
{ | *{{TAG|FINDIFF}}=1 Use a forward ({{TAG|FINDIFF}}=1) or central ({{TAG|FINDIFF}}=2) difference formula for the numerical differentiation to compute the curvature along the dimer direction | ||
*{{TAG|DIMER_DIST}}=0.01 The step size for a numerical differentiation (in <math>\AA</math>) | |||
*{{TAG|MINROT}}=0.01 Dimer is rotated only if the predicted rotation angle is greater than {{TAG|MINROT}} (rad.) | |||
*{{TAG|STEP_SIZE}}=0.01 Trial step size for optimization step (in <math>\AA</math>) | |||
*{{TAG|STEP_MAX}}=0.1 Trust radius (upper limit) for the optimization step (in <math>\AA</math>) | |||
Important information about the progress of optimization is written in the {{FILE|OUTCAR}} file after the expression ''DIMER METHOD''. | |||
{ | |||
In particular, it is useful to check the curvature along the dimer direction, which should be a negative number (a long sequence of positive numbers usually indicates that the algorithm fails to converge to the correct transition state). | |||
{ | {{NB|mind| The current implementation does not support lattice optimizations ({{TAG|ISIF}}>2) and can be used only for the relaxation of atomic positions.}} | ||
== Initial dimer axis == | |||
The direction of an unstable vibrational mode can be obtained by performing the vibrational analysis ({{TAG|IBRION}}=5) and taking the x-, y-, and z- components of the imaginary vibrational mode (after division by <math>\sqrt{m}</math>!) parallel with the reaction coordinate. Note that in order to plot "Eigenvectors after division by SQRT(mass)", {{TAG|NWRITE}}=3 should be used. | |||
{ | |||
== Practical example == | |||
In this example, the transition state for the ammonia flipping is computed. All calculations discussed here were performed using the PBE functional, Brillouin zone sampling was restricted to the gamma point. This practical example can be completed in a few seconds on a standard desktop PC. The starting structure for the IDM simulation should be a reasonable guess for the transition state. A {{FILE|POSCAR}} file with the initial guess for the ammonia flipping looks like this: | |||
in the file | |||
ammonia flipping | |||
1. | |||
6. 0. 0. | |||
0. 7. 0. | |||
0. 0. 8. | |||
H N | |||
3 1 | |||
cart | |||
-0.872954 0.000000 -0.504000 | |||
0.000000 0.000000 1.008000 | |||
0.872954 0.000000 -0.504000 | |||
0.000000 0.000000 0.000000 | |||
As an input for the dimer method, the direction of the unstable mode (dimer axis) is needed. This can be obtained by performing vibrational analysis. The {{FILE|INCAR}} file should contain the following lines: | |||
{{TAGBL|NSW}} = 1 | |||
{{TAGBL|Prec}} = Normal | |||
{{TAGBL|IBRION}} = 5 ! perform vibrational analysis | |||
{{TAGBL|NFREE}} = 2 ! select central differences algorithm | |||
{{TAGBL|POTIM}} = 0.02 ! step for the numerical differenciation | |||
{{TAGBL|NWRITE}} = 3 ! write down eigenvectors of dynamical matrix after division by SQRT(mass) | |||
After completing the vibrational analysis, we look up the hardest imaginary mode (Eigenvectors after division by SQRT(mass)!) in the {{FILE|OUTCAR}} file: | |||
12 f/i= 23.224372 THz 145.923033 2PiTHz 774.681641 cm-1 96.048317 meV | |||
X Y Z dx dy dz | |||
5.127046 0.000000 7.496000 0.000001 0.522103 -0.000009 | |||
0.000000 0.000000 1.008000 -0.000006 0.530068 0.000000 | |||
0.872954 0.000000 7.496000 -0.000005 0.522067 -0.000007 | |||
0.000000 0.000000 0.000000 0.000001 -0.111442 0.000001 | |||
and use the last three columns to define the dimer axis in the {{FILE|POSCAR}} file: | |||
ammonia flipping | |||
1. | |||
6. 0. 0. | |||
0. 7. 0. | |||
0. 0. 8. | |||
H N | |||
3 1 | |||
cart | |||
-0.872954 0.000000 -0.504000 ! coordinates for atom 1 | |||
0.000000 0.000000 1.008000 | |||
0.872954 0.000000 -0.504000 | |||
0.000000 0.000000 0.000000 ! coordinates for atom N | |||
! here we define trial unstable direction: | |||
0.000001 0.522103 -0.000009 ! components for atom 1 | |||
-0.000006 0.530068 0.000000 | |||
-0.000005 0.522067 -0.000007 | |||
0.000001 -0.111442 0.000001 ! components for atom N | |||
In order to perform IDM calculation, the {{FILE|INCAR}} file should contain the following lines: | |||
{{TAGBL|NSW}} = 100 | |||
{{TAGBL|Prec}}=Normal | |||
{{TAGBL|IBRION}}=44 ! use the dimer method as optimization engine | |||
{{TAGBL|EDIFFG}}=-0.03 | |||
With this setting, the algorithm converges in just a few relaxation steps. Further vibrational analysis can be performed to prove that the relaxed structure is indeed a first-order saddle point (one imaginary frequency). | |||
- | |||
{{NB|mind| | |||
If you perform a frequency calculation on this optimized structure, take care to consider the translational modes that are present. These will appear as small imaginary frequencies, less than 5 cm-1 in the OUTCAR file and may be recognized by the "dx" terms for each atom being large, while the remaining "dy" and "dz" terms are near zero (for x translation). This will result in multiple imaginary frequencies. The goal is to have one "large" imaginary frequency, and then these small translational modes should be set to zero for calculating thermodynamic properties. | |||
}} | |||
For example, the x-translation is shown below: | |||
- | |||
11 f/i= 0.092375 THz 0.580407 2PiTHz 3.081284 cm-1 0.382030 meV | |||
X Y Z '''dx''' dy dz | |||
5.127046 0.000000 7.496000 '''-0.247337''' 0.000339 0.008273 | |||
0.000000 0.000000 1.008000 '''-0.232726''' 0.000336 -0.000038 | |||
0.872954 0.000000 7.496000 '''-0.247322''' 0.000340 -0.008357 | |||
0.000000 0.000000 0.000000 '''-0.242501''' 0.000341 -0.000041 | |||
== Related tags and articles == | |||
{{TAG|FINDIFF}}, {{TAG|DIMER_DIST}}, {{TAG|MINROT}}, {{TAG|STEP_SIZE}}, {{TAG|STEP_MAX}} | |||
== References == | == References == | ||
---- | ---- | ||
[[Category: | [[Category:Transition states]][[Category:Ionic minimization]][[Category:Howto]] | ||
Latest revision as of 10:52, 28 October 2024
The dimer method[1] is a technique for the optimization of transition states. In VASP, the improved dimer method (IDM) by Heyden et al. is implemented, and a detailed presentation of the method can be found in their paper[2].
The initial curvature along the dimer axis is computed using finite differences. The initial dimer direction must be provided (see below). The IDM procedure shown in Figure 1 is described in four steps with FINDIFF = 1:
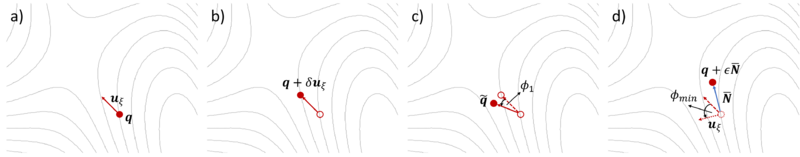
a) An initial direction uξ is taken from the most negative vibrational mode of the trial structure. The trial structure is the first point q.
b) An additional point on the potential energy surface (PES) forward along the trial direction is defined q + δuξ. The first and second points together define the dimer.
c) The dimer is then rotated on the PES about q by angle φ1 to q̃ such that its axis is parallel to the direction of maximal negative curvature.
d) A new direction is defined by rotating uξ by φmin to minimize the negative curvature of the PES λ. A search direction N̅ is defined using a minimization algorithm, then a translation (optimization step) is taken in the unstable direction of N̅ to q + εN̅, where ε is a step distance. The potential energy is is maximized along the unstable direction, (i.e., dimer axis) while it is minimized in all other directions.
Rotation followed by translation is followed iteratively until convergence, i.e. the saddle point, is reached.
The method is invoked by setting IBRION=44 in the INCAR file.
Furthermore, the user must specify the direction of the unstable mode. The corresponding dimensional vector is defined in the POSCAR file after the lines with atomic coordinates and a separating blank line. Note that the dimer direction is automatically normalized, i.e., the norm of the dimer axis is irrelevant. An example of a POSCAR file for a simulation with the dimer method is given in the following:
ammonia flipping
1.
6. 0. 0.
0. 7. 0.
0. 0. 8.
H N
3 1
cart
-0.872954 0.000000 -0.504000 ! coordinates for atom 1
0.000000 0.000000 1.008000
0.872954 0.000000 -0.504000
0.000000 0.000000 0.000000 ! coordinates for atom N
! here we define trial unstable direction:
0.000001 0.522103 -0.000009 ! components for atom 1
-0.000006 0.530068 0.000000
-0.000005 0.522067 -0.000007
0.000001 -0.111442 0.000001 ! components for atom N
As in the other structural optimization algorithms in VASP, convergence is controlled through the EDIFFG tag.
Experienced users can affect the performance of the dimer method by modifying the numerical values of the following parameters (the given example values are the default values):
- FINDIFF=1 Use a forward (FINDIFF=1) or central (FINDIFF=2) difference formula for the numerical differentiation to compute the curvature along the dimer direction
- DIMER_DIST=0.01 The step size for a numerical differentiation (in )
- MINROT=0.01 Dimer is rotated only if the predicted rotation angle is greater than MINROT (rad.)
- STEP_SIZE=0.01 Trial step size for optimization step (in )
- STEP_MAX=0.1 Trust radius (upper limit) for the optimization step (in )
Important information about the progress of optimization is written in the OUTCAR file after the expression DIMER METHOD.
In particular, it is useful to check the curvature along the dimer direction, which should be a negative number (a long sequence of positive numbers usually indicates that the algorithm fails to converge to the correct transition state).
| Mind: The current implementation does not support lattice optimizations (ISIF>2) and can be used only for the relaxation of atomic positions. |
Initial dimer axis
The direction of an unstable vibrational mode can be obtained by performing the vibrational analysis (IBRION=5) and taking the x-, y-, and z- components of the imaginary vibrational mode (after division by !) parallel with the reaction coordinate. Note that in order to plot "Eigenvectors after division by SQRT(mass)", NWRITE=3 should be used.
Practical example
In this example, the transition state for the ammonia flipping is computed. All calculations discussed here were performed using the PBE functional, Brillouin zone sampling was restricted to the gamma point. This practical example can be completed in a few seconds on a standard desktop PC. The starting structure for the IDM simulation should be a reasonable guess for the transition state. A POSCAR file with the initial guess for the ammonia flipping looks like this:
ammonia flipping
1.
6. 0. 0.
0. 7. 0.
0. 0. 8.
H N
3 1
cart
-0.872954 0.000000 -0.504000
0.000000 0.000000 1.008000
0.872954 0.000000 -0.504000
0.000000 0.000000 0.000000
As an input for the dimer method, the direction of the unstable mode (dimer axis) is needed. This can be obtained by performing vibrational analysis. The INCAR file should contain the following lines:
NSW = 1 Prec = Normal IBRION = 5 ! perform vibrational analysis NFREE = 2 ! select central differences algorithm POTIM = 0.02 ! step for the numerical differenciation NWRITE = 3 ! write down eigenvectors of dynamical matrix after division by SQRT(mass)
After completing the vibrational analysis, we look up the hardest imaginary mode (Eigenvectors after division by SQRT(mass)!) in the OUTCAR file:
12 f/i= 23.224372 THz 145.923033 2PiTHz 774.681641 cm-1 96.048317 meV
X Y Z dx dy dz
5.127046 0.000000 7.496000 0.000001 0.522103 -0.000009
0.000000 0.000000 1.008000 -0.000006 0.530068 0.000000
0.872954 0.000000 7.496000 -0.000005 0.522067 -0.000007
0.000000 0.000000 0.000000 0.000001 -0.111442 0.000001
and use the last three columns to define the dimer axis in the POSCAR file:
ammonia flipping
1.
6. 0. 0.
0. 7. 0.
0. 0. 8.
H N
3 1
cart
-0.872954 0.000000 -0.504000 ! coordinates for atom 1
0.000000 0.000000 1.008000
0.872954 0.000000 -0.504000
0.000000 0.000000 0.000000 ! coordinates for atom N
! here we define trial unstable direction:
0.000001 0.522103 -0.000009 ! components for atom 1
-0.000006 0.530068 0.000000
-0.000005 0.522067 -0.000007
0.000001 -0.111442 0.000001 ! components for atom N
In order to perform IDM calculation, the INCAR file should contain the following lines:
NSW = 100 Prec=Normal IBRION=44 ! use the dimer method as optimization engine EDIFFG=-0.03
With this setting, the algorithm converges in just a few relaxation steps. Further vibrational analysis can be performed to prove that the relaxed structure is indeed a first-order saddle point (one imaginary frequency).
| Mind:
If you perform a frequency calculation on this optimized structure, take care to consider the translational modes that are present. These will appear as small imaginary frequencies, less than 5 cm-1 in the OUTCAR file and may be recognized by the "dx" terms for each atom being large, while the remaining "dy" and "dz" terms are near zero (for x translation). This will result in multiple imaginary frequencies. The goal is to have one "large" imaginary frequency, and then these small translational modes should be set to zero for calculating thermodynamic properties. |
For example, the x-translation is shown below:
11 f/i= 0.092375 THz 0.580407 2PiTHz 3.081284 cm-1 0.382030 meV
X Y Z dx dy dz
5.127046 0.000000 7.496000 -0.247337 0.000339 0.008273
0.000000 0.000000 1.008000 -0.232726 0.000336 -0.000038
0.872954 0.000000 7.496000 -0.247322 0.000340 -0.008357
0.000000 0.000000 0.000000 -0.242501 0.000341 -0.000041
Related tags and articles
FINDIFF, DIMER_DIST, MINROT, STEP_SIZE, STEP_MAX
References
- ↑ G. Henkelman and H. Jónsson, A dimer method for finding saddle points on high dimensional potential surfaces using only first derivatives, J. Chem. Phys. 111, 7010–7022 (1999).
- ↑ A. Heyden, A. T. Bell, and F. J. Keil, Efficient methods for finding transition states in chemical reactions: Comparison of improved dimer method and partitioned rational function optimization method, J. Chem. Phys. 123, 224101 (2005).
